Let us help you select the appropriate Sonicator model and accessory for your application. Call us at 203.426.0101 or
fill out a Contact Form.
fill out a Contact Form.
×
Qsonica offers several devices routinely used for shearing chromatin and DNA. The Q800R system is our most advanced option and offers the highest throughput. In addition, the Q700 and Q125 are very popular and effective alternatives.

For individual samples the easiest method is to insert a small probe directly into a sample. At Qsonica, we call this direct probe sonication. Energy is transmitted from the probe directly into the sample and the entire volume is processed very quickly.
Indirect sonication is a great option for processing very small samples as well as multiple samples. Contact a Qsonica representative to find out which option is best for you.
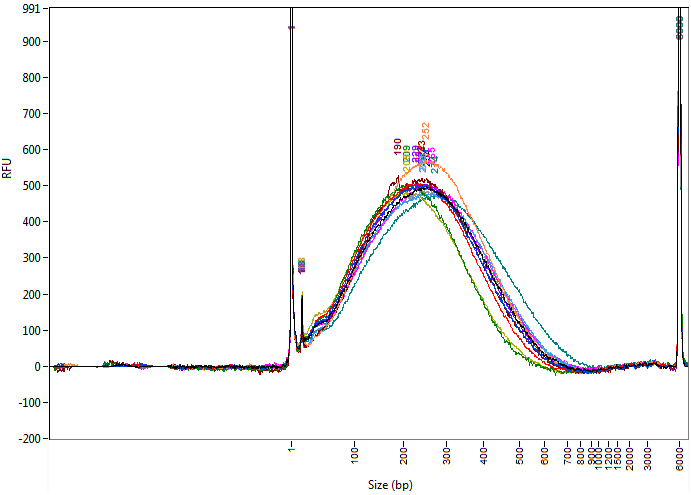

The Q800R3 is our 2nd generation DNA and chromatin shearing system. Improvements include an enhanced user-friendly design and quieter operation while remaining thousands of dollars less than the competition.
The system is compatible with commercially available sample tubes and is capable of processing up to 18 samples at one time.
Find out why customers like NIH, Harvard and Genentech have adopted our technology for use with multiple NGS platforms and see the new Illumina® technical note highlighting the Q800R3 system.
“We’ve been using Qsonica Q800R in our NGS library preparation pipeline for more than two years. Over that time, we processed hundreds of samples and Q800R has proven to be an exceptionally reliable machine producing consistent results with little variability from run to run. Q800R allows us to finely control the size distribution of sheared fragments. Its ability to process up to 18 samples at a time provides more than sufficient capacity for our workflow.”
Museum Genomics Illuminate the High Specificity of a Bioluminescent Symbiosis for a Genus of Reef Fish
Alison L. Gould, Allison Fritts-Penniman and Ana Gaisiner, Frontiers in Ecology and Evolution, Feb. 2021.
Isolation of extracellular vesicles from intestinal tissue in a mouse model of intestinal ischemia/reperfusion injury
Xiao-Dong Chen, Jin Zhao, et al., Biotechniques, 2020.
Adsorption of double-stranded ribonucleic acids (dsRNA) to iron (oxyhydr-) oxide surfaces: comparative analysis of model dsRNA molecules and deoxyribonucleic acids (DNA)
Katharina Sodnikar, Kimberly M. Parker, et al., Royal Society of Chemistry, 2021
More than 50% of Clostridium difficile Isolates from Pet Dogs in Flagstaff, USA, Carry Toxigenic Genotypes
Stone, N.E., et al., PLOS ONE | DOI:10.1371/journal.pone.0164504 October 10, 2016
The complete mitochondrial genome of Cerion uva uva (Gastropoda: Panpulmonata: Stylommatophora: Cerionidae)
Harasewych, M. G. et al., Mitchondrial DNA Part B: Resources, 2017 VOL. 2, NO. 1, pp 159–160, DOI: 10.1080/23802359.2017.1303343
Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis
Xindan Wang, Tung B.K. Le, Bryan R. Lajoie, et al., Genes & Dev. 2015 29: pp 1661-1675, Cold Spring Harbor Laboratory Press
Phylogenomics, biogeography and diversification of obligate mealy bug tending ants in the genus Acropyga
Blaimer et al., Molecular Phylogenetics and Evolution, Vol. 102, September 2016, pp 20-29
Sequence Capture and Phylogenetic Utility of Genomic Ultraconserved Elements Obtained from Pinned Insect Specimens
Bonnie B. Blaimer, et. al., PLoS ONE 11(8):e0161531.doi:10.137
An RNF168 fragment defective for focal accumulation at DNA damage is proficient for inhibition of homologous recombination in BRCA1 deficient cells
Munoz, et. al., 2014; Nucleic Acids Research, doi: 10.1093/nar/gku421
Enhanced virome sequencing using targeted sequence capture
Wylie, et. al., 2016; Genome Research, 25: 1910-1920
Phylogenetic analysis of higher-level relationships within Hydroidolina (Cnidaria: Hydrozoa) Using mitochondrial genome data and insight into their mitochondrial transcription
Kayal, et. al., 2015; PeerJ, 3: e1403; doi 10.7717/peerj.1403
Revisiting the Zingiberales: using multiplexed exon capture to resolve ancient and recent phylogenetic splits in a charismatic plant lineage
Sass, et. al., 2016; PeerJ, 4:e1584; doi 10.7717/peerj.1584
CXCL8 histone H3 acetylation is dysfunctional in airway smooth muscle in asthma: regulation by BET
Clifford, et. al., Am J Physiol Lung Cell Mol Physiol Vol. 308: L962–L972, February 2015
IƙB Kinase β (IKBKB) Mutations in Lymphomas That Constitutively Activate Canonical Nuclear Factor ƙB (NFƙB) Signaling*
Kai, et. al., Journal of Biological Chemistry Vol. 289, No. 38, pp26960-26972, September 2014
Farnesoid X receptor regulates forkhead Box O3a activation in ethanol-induced autophagy and hepatotoxicity
Manley, et. al., Redox Biology Vol. 2, pp991-1002, August 2014
Inhibition of mutant EGFR in lung cancer cells triggers SOX2-FOXO6-dependent Survival pathways
Rothenberg, et. al., eLife, 2015; 4:e06132. DOI: 10.7554/eLife.
DNA Damage Response Factors from Diverse Pathways, Including DNA Crosslink Repair, Mediate Alternative End Joining
Howard, et. al., PLOS Genetics, 11(1): e1004943, 2015; doi:10.1371/journal.pgen.1004943
KDM4A Lysine Demethylase Induces Site-Specific Copy Gain and Rereplication of Regions Amplified in Tumors
Black et.al., Cell, Volume 154, Issue 3, 541-555, 18 July 2013
Enhanced in vivo Fitness of Carbapenem-resistant oprD mutants of Pseudomonas Aeruginosa Revealed Through High-throughput Sequencing
Skurnika et al., PNAS, Volume 110 No. 51, 20747–20752, 17 December 2013
Coupling Mutagenesis and Parallel Deep Sequencing to Probe Essential Residues in a Genome or Gene
Robins et al., PNAS, Volume 110 No. 9, E848–E857, 26 February 2013
TP53 supports basal-like differentiation of mammary epithelial cells by preventing translocation of deltaNp63 into nucleoli
Munne, P.M. et.al., Nature Sci. Rep., 4, 4663; DOI: 10.1038/srep04663 (2014)
Rediscovery of an Endemic Vertebrate from the Remote Islas Revillagigedo in the Eastern Pacific Ocean: The Clarion Nightsnake Lost and Found
Mulcahy et al., PLOS ONE, Volume 9 Issue No. 5, E97682, 2 May 2015
MELK is an oncogenic kinase essential for mitotic progression in basal-like breast cancer cells Wang et al., eLife 2014; 3:e01763. DOI:10.7554/eLife.01763
SH2B1 in _-Cells Promotes Insulin Expression and Glucose Metabolism in Mice
Chen, et. al., Molecular Endocrinology, Volume 28, Issue 5, 696-705, May 2014
O-GlcNAcase expression is sensitive to changes in O-GlcNAc homeostasis
Zhang, et. al., Frontiers in Endocrinology, Volume 5, Article 206, December 2014 http://dx.doi.org/10.3389/fendo.2014.00206
Lysine Demethylase KDM4A Associates with Translation Machinery and Regulates Protein Synthesis
Van Rechem et.al., AACR Cancer Discovery 2015, Volume 5, 255-263 DOI: 10.1158/2159-8290.CD-14-1326
Target enrichment of ultraconserved elements from arthropods provides a genomic perspective on relationships among Hymenoptera
Faircloth et.al., Molecular Ecology Resources, Volume 15, 489-201, 2015
SOX4 enables oncogenic survival signals in acute lymphoblastic leukemia
Ramezani-Rad et.al., Blood Journal, Volume 121 (1), 1-9, October 2012
ZASC1 Stimulates HIV-1 Transcription Elongation by Recruiting P-TEFb and TAT to the LTR Promoter
Bruce et.al., PLOS Pathogens, Volume 9, Issue 10, 1-19, October 2013
Activation of TGF-b1 Promoter by Hepatitis C Virus-Induced AP-1 and Sp1: Role of TGF-b1 in Hepatic Stellate Cell Activation and Invasion
Presser et.al., PLOS ONE, Volume 8, Issue 2, 1-19, February 2013
Allelic Imbalance Assays to Quantify Allele-Specifi c Gene Expression and Transcription Factor Binding
Luca et.al., Pharmacogenomics: Methods and Protocols, Methods in Molecular Biology, vol. 1015, DOI 10.1007/978-1-62703-435-7_13, 201-211, (2013)
Cdk1 Regulates the Temporal Recruitment of Telomerase and Cdc13-Stn1-Ten1 Complex for Telomere Replication
Liu et.al., Molecular and Cellular Biology, Volume 34, Number 1, 57-70 , January 2014
Wnt Signaling Inhibits Adrenal Steroidogenesis by Cell-Autonomous and Non–Cell-Autonomous Mechanisms
Walczak et. al., Molecular Endocrinology, Volume 28, No. 9, pp 1471-1486; September 2014
Tissue-specific regulation of Igf2r/Airn imprinting during gastrulation
Marcho et. al., Epigenetics and Chromatin, Volume 8, No.10, 2015; DOI 10.1186/s13072-015-0003-y
Parasitology Canonical histone H2Ba and H2A.X dimerize in an opposite genomiclocalization to H2A.Z/H2B.Z dimers in Toxoplasma gondii
Bogado et.al., Molecular and Biochemical Parasitology, 197 (2014) 36-42
Genome-wide Mapping and Characterization of Notch-Regulated Long Noncoding RNAs in Acute Leukemia
Trimarchi et.al., Cell, Volume 158, 593-606, July 2014
The capacity of target silencing by Drosophila PIWI and piRNAs
Post et.al., Cold Spring Harbor Laboratory Press, RMA Volume 20: 1977-1986, April 2015
Chromatin Immunoprecipitation with Fixed Animal Tissues and Preparation for High-Throughput Sequencing
Cotney et.al., Cold Spring Harbor Laboratory Press (2015)
Cold Spring Harb Protoc; doi: 10.1101/pdb.prot084848